Breaking the Big Data Bottleneck: Solving Spark’s “Small Files” Problem
How DataFlint helps you identify and fix small-file overhead for faster Spark performance
The first time I (Meni) developed a big data application with Apache Spark my spark job couldn’t finish because I partitioned the data incorrectly and accidentally wrote millions of extremely small files to S3.
As time went on I saw that this kind of development experience was very common in the big data world.
I learned to use tools like Spark UI but also discovered it’s (many) limitations, mainly it’s just not very human readable[1].
A Real World Example
Let’s try to recreate this exact scenario with a simple PySpark script:
from pyspark.sql import SparkSession
spark = SparkSession \
.builder \
.appName("Sales Filterer") \
.master("local[1]") \
.getOrCreate()
df = spark.read.load("~/data/store_sales")
df_filtered = df.filter(df.ss_quantity > 1)
df_filtered.write \
.mode("overwrite") \
.partitionBy("ss_quantity") \
.parquet("/tmp/store_sales")
spark.stop()In this simple example we read some sales related data as we done in previous post[1], we filter by items that have quantity bigger than 1 and then saving the data partitioned by the quantity[2].
Even though the code might seem straightforward, on my machine[3] it takes over a minute to run for a dataset of only ~273 MB!
Why Is It Running So Slowly? The Small Files Problem
When you run into a situation where your dataset is split into lots of tiny files (for example, thousands or millions of files in the KB range), Spark or any other Big Data engine will spend most of its time handling overhead rather than actual data processing.
A few reasons why these “small files” cause poor performance:
Excessive Task Creation
Big data engines typically create a task or map task for each file or block. When you have hundreds or thousands of small files, the engine needs to spin up a large number of tasks just to read them, adding scheduling, I/O, and resource overhead.Metadata Overhead
Each file (no matter how small) has associated metadata (file name, permissions, location, etc.) that has to be fetched and processed. Doing this for many files greatly amplifies the time spent looking up and parsing metadata instead of doing actual work.I/O Overhead
Each small file requires establishing a connection or reading a file header and footer. With remote storage systems like HDFS or S3, each read operation can have a notable startup cost. So a thousand 100 KB files can be worse than a single 100 MB file because the engine has to continuously renegotiate I/O connections.Worsened Data Skew
Spark tries to distribute data across executors, but if there are many tiny files, some executors might handle more files (i.e., more overhead) than others, leading to skew and inefficiency.Inefficient Compression / Merging
File formats like Parquet are optimized for larger blocks. Tiny files can negate some of these optimizations, forcing Spark to repeatedly initialize readers and compress smaller chunks less effectively.
In our specific example, each small file is read from HDFS, filtered, and then re-partitioned and written back out. Because the files are already tiny, Spark ends up generating even smaller files during the write stage:
Reading small files + partitioning = writing even smaller files
Performance boost: Enter DataFlint
DataFlint is an open source performance monitoring library for Apache Spark, developed by Meni Shmueli, aimed at identifying these exact sorts of problems.
To install it all we need to do is to add this 2 lines:
.config("spark.jars.packages", "io.dataflint:spark_2.12:0.2.9") \
.config("spark.plugins", "io.dataflint.spark.SparkDataflintPlugin") \To our python application:
from pyspark.sql import SparkSession
spark = SparkSession \
.builder \
.appName("Sales Filterer") \
.config("spark.jars.packages", "io.dataflint:spark_2.12:0.2.9") \
.config("spark.plugins", "io.dataflint.spark.SparkDataflintPlugin") \
.master("local[1]") \
.getOrCreate()
df = spark.read.load("~/data/store_sales")
df_filtered = df.filter(df.ss_quantity > 1)
df_filtered.write \
.mode("overwrite") \
.partitionBy("ss_quantity") \
.parquet("/tmp/store_sales")
spark.stop()(for more installation options, see the docs)
Now when we will open Spark UI we will have a new button that will open DataFlint
Identifying the performance issue
Once we enter DataFlint for our spark job, we can see that DataFlint identified a “reading small files” issue in real-time during the query run
And when the query ends we can that DataFlint also identified a “writing small partitioned files” issue:
And finally this really simple query of filtering only ~273MB of data, taking 51 seconds to be completed
So what exactly is going on? and how can we fix it?
Small files IO in big data
Big Data engines are usually optimized to work with files in size ranges of 128MB-1GB, working with really small files such as in our case (DataFlint calculated average of 85KB per file we read) can cause all sorts of performance problems, some are not so obvious than others.
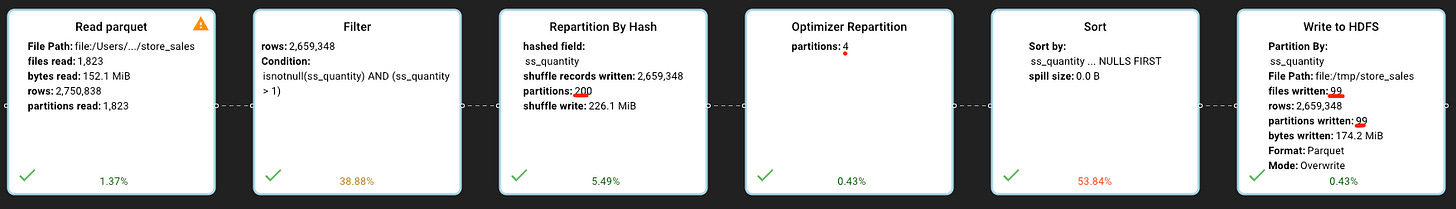
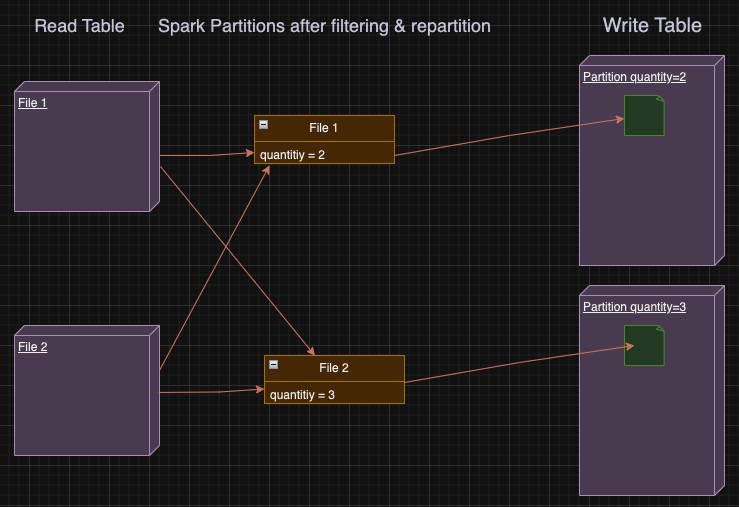
In our case, for each small file spark reads from HDFS, Spark will apply the filter, partition the remaining data by quantity and save each partition of the small file to HDFS, resulting on writing even smaller files!
How can we fix it?
Fixing the writing small files issue
In this scenario we will focus on fixing the writing small files problem, as our source table file sizes might not be in our control.
Luckily for us, DataFlint identified a “writing small partitioned files” issue for us and even suggesting a fix:
Telling spark to repartition the data in-memory by our table partition key:
Now let’s apply this fix in our code:
from pyspark.sql import SparkSession
spark = SparkSession \
.builder \
.appName("Sales Filterer") \
.config("spark.jars.packages", "io.dataflint:spark_2.12:0.2.7") \
.config("spark.plugins", "io.dataflint.spark.SparkDataflintPlugin") \
.master("local[1]") \
.getOrCreate()
df = spark.read.load("~/data/store_sales")
df_filtered = df.filter(df.ss_quantity > 1)
df_repartitioned = df_filtered.repartition("ss_quantity") = # THE FIX
df_repartitioned.write \
.mode("overwrite") \
.partitionBy("ss_quantity") \
.parquet("/tmp/store_sales")
spark.stop()And see the updated query plan in DataFlint:
We can see that our query has 2 new steps — repartition by quantity hash to 200 partitions (200 is a spark default for repartitioning), and a spark optimizer repartitioned it to 4 partitions.
By hash partitioning by quantity we ensure that each spark partition has all the necessary data for writing exactly 1 file to that table’s partition — meaning having all the records with the same quantity value.
Now the fixed query is only taking only 15 seconds instead of 51 seconds, more than 300% speed boost!
Final Thoughts
Small file issues are pervasive in big data. Big Data engines are optimized for fewer, larger files (128 MB–1 GB range). When your data ends up in lots of tiny files, overhead, task creation, I/O, metadata becomes the bottleneck.
Tools like DataFlint help you quickly pinpoint these bottlenecks, offer actionable fixes, and make your Spark development process more efficient.
If you’d like to try DataFlint yourself, check out the DataFlint GitHub repository.
We’d love to hear your thoughts and ideas, if you have something to contribute, feel free to join the conversation and make an impact!
Notes
[1] Check out our privous blog on naming spark jobs:
Make spark jobs readable again!
Your go to resource for actionable insights into big data performance. Each week, we’ll share tips, tools, and techniques to help you optimize your workflows. If you’re dealing with large datasets, distributed systems, and are tired of fluff, this newsletter is for you!
[2] The choice of the partition key for this example arbitrary, in the case of TPC-DS the quantity field has low cardinality so it make sense to partition by it for this example.
[3] 2023 16 inch MacBook pro, if running on all cores it runs too fast to be able to see the Spark/dataflint UI so I added a local[1] configuration to limit spark to use only 1 core.
[4] in addition, this performance problem can accur without reading small files. For example, if you read a lot of big files but most of the data has been filtered